DIFF_CALC Loading
After importing the data, it is possible to immediately use DIFF_PREP for image registration and artifact correction. However, it is advisable, particularly if you are new to this software, to load your data into DIFF_CALC and inspect the raw data, then perform a simple linear tensor fitting to make sure that the b-matrix is correct.
- Start DIFF_CALC following the instructions.
- Click on load images and select your listfile.
- To inspect the raw data, click on display raw images and use the SLICES and B_VALUES sliders to look through the volumes and slices of your data. This is shown in detail in Display Raw Images
- To do a quick linear tensor fit
- First click on Estimate signal SD to estimate the image noise by either drawing an ROI in the background, or specifying an a priori value
- Then click on Mask raw images to calculate a brain mask
- Then select the "opt" button to the right of the process tensor button and select the linear fitting option
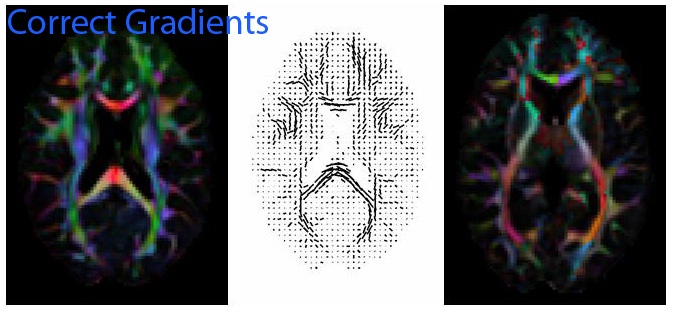
- When the fit is complete, click on roi utilities, and inspect the linefield and the DEC maps in order to assure you have a correct b-matrix. The figures below show correct and incorrect linefield and DEC maps for one dataset.
The following figure shows some images of a tensor fit with the y-gradient flipped incorrectly. From left to right 1) absolute value DEC map. It looks reasonable. 2) The Linefield image—note the vertical tensor direction in the splenium of the corpus callosum. This indicates there is a problem with the gradient file provided. 3) The no symmetry DEC map on a lower slice in the brain—note the pink and orange colors in the internal capsule. This is incorrect.

The following figure shows the same slices as the previous figure, but with the y gradient flipped. Note the continuous left-right tensor directions in the corpus callosum on the linefield image, and the correct green and blue colors in the internal capsule.
Please click on the linked pages to get more details about the above steps.
