Phantom Analysis Utility
The Phantom Analysis utility implements the two-step analysis framework for multi-center DTI studies as described in our recent publication in Human Brain Mapping.1
Two-step analysis framework
Step 1: Voxel-wise median image is computed from all input images. Difference images are computed for each timepoint from the median image.
Step 2: assessment of overall variance, similar to the definition of the traditional ANOVA, compute the intrasite (i.e., within site) and the intersite (i.e., between sites) variances. In addition, we compute the variability (i.e. standard deviation) maps, and ICC coefficients.
Preparing your phantom data for analysis:
- Pre-process your data using DIFF_PREP, including correction for motion, eddy current distortion and EPI distortion.
- Use a common target image for the corrections, a good choice might be a T2W image from the 1st timepoint of the phantom. Using a common target is important, as this analysis is performed on a voxel-wise basis, and assumes that the input images are decently co-registered.
- Compute the tensor and tensor derived metrics using DIFF_CALC.
- Create a text file listing the full path to all site and all time point images. An example text file with appropriate format is shown below.
- The analysis should be run, at a minimum, on fractional anisotropy (FA) and trace of the diffusion tensor (TR). It can be run on any tensor derived metric. Interpretation of the results may be difficult for metrics that include negative numbers, such as skewness, and should be used with caution on this type of metric.
To run the software:
First, navigate to TORTOISE_V1.3.0/Utilities/Phantom_Analysis, then do one of the following options:
- Double click on varvm
- Type varvm (or ./varvm) at the command line
- If IDL VM is installed: type idl -vm=var_gui.sav at the command line
- If IDL full version is installed: start IDL, then type var_gui at the IDL command prompt
Click on the splash screen, and a GUI will pop up that looks like:
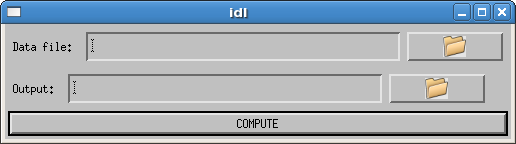
To run the software:
- Use the browse button (at right) to input your data file. The data file is a text file that lists the number of timepoints at each site on consecutive lines, followed by the full path to each image file in the same order.
- Example: Two sites, Site 1 has 4 timepoints, and Site 2 has 3 timepoints.
- Make sure there are no blank lines at the end of the text file, or the software may crash
4
3
/data/site1_timepoint1_proc/site1_timepoint1_N1_SAVE/site1_timepoint1_FA.nii
/data/site1_timepoint2_proc/site1_timepoint2_N1_SAVE/site1_timepoint2_FA.nii
/data/site1_timepoint3_proc/site1_timepoint3_N1_SAVE/site1_timepoint3_FA.nii
/data/site1_timepoint4_proc/site1_timepoint4_N1_SAVE/site1_timepoint4_FA.nii
/data/site2_timepoint1_proc/site2_timepoint1_N1_SAVE/site2_timepoint1_FA.nii
/data/site2_timepoint2_proc/site2_timepoint2_N1_SAVE/site2_timepoint2_FA.nii
/data/site2_timepoint3_proc/site2_timepoint3_N1_SAVE/site2_timepoint3_FA.nii
- Use the browse button (at right) to provide your desired output directory.
- Click COMPUTE
Outputs of the software:
In the provided output directory, there will now be a subdirectory called outlier_analysis, and 7 nifti images.
- outlier_analysis folder contains the difference from median image for each input image.
- example: diff_site1_timepoint1_FA.nii, diff_site1_timepoint2_FA.nii, etc.
- icc_inter.nii - between sites ICC
- icc_intra.nii - within sites ICC
- intersite_variability.nii - between sites variability
- intersite_variance.nii - between sites variance
- intrasite_variability.nii - within sites variability
- intrasite_variance.nii - within sites variance
- mean_img.nii - mean image computed from all input images
Formulas for the various quantities are defined in Walker, et. al.1
References
- Walker, L., Curry, M., Nayak, A., Lange, N., Pierpaoli, C., and the Brain Development Cooperative Group, A Framework for the Analysis of Phantom Data in Multicenter Diffusion Tensor Imaging Studies, Human Brain Mapping, 2012, PMID:22461391
