Transformation Files
Description of the transformation files in TORTOISE (v1.0.5)
After running the full diffprep pipeline there should be two transformation files: ..._rpd.transformations and ..._DMC.transformations. The rpd transformation file contains information about the transformations for the eddy distortion correction and the motion correction. The DMC transformation file contains information about the final rigid reorientation to the structural target image.
Things to know
- Number of rows in the transformation file = number of volumes in your dataset
- A volume is defined as the set of images making up the whole brain for a single b0 or DWI measurement
- The 1st b0 (i.e. first row if your first volume is a b0 image) is treated slightly differently than the rest of the volumes.
- First row: b0 → structural: If rigid only, this will be 6 parameters, the rest of the columns will be either 0 or 1. If eddy distortion correction, there will be values for all columns, as described below.
- All subsequent rows: DWI (or subsequent b0) → first b0: the transformation for each row is applied to its volume, followed by the transformation from the first row.
rpd.transformations
1st set of three columns represents the rigid portion of the transformation (in millimeters)
Column #1 > x (for axial data = RL)
Column #2 > y (for axial data = AP)
Column #3 > z (for axial data = IS)
2nd set of three columns represents the rotational (Euler angles) component of the transformation (in radians)
Column #4 > θx
Column #5 > θy
Column #6 > θz
where
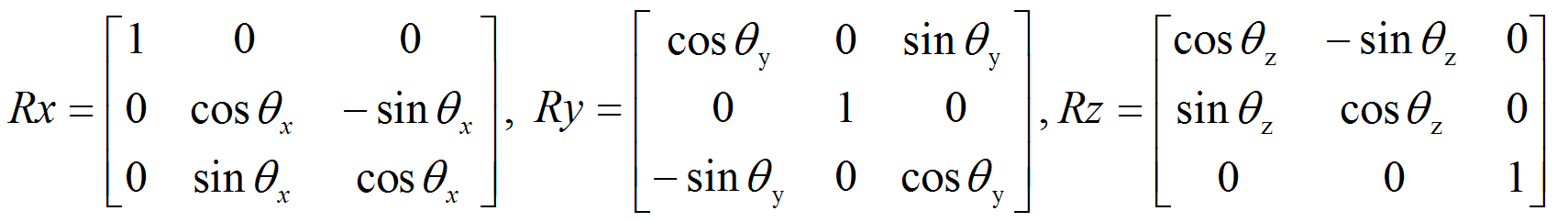
And overall rotation R= Rx Ry Rz
3rd set of three columns represents the weights of the linear affine component of the transformation (shears) (unitless)
Example: if no eddy correction is applied, and you have a horizontal phase encode direction, these values would be x=1, y,z=0
Column #7 > x
Column #8 > y
Column #9 > z
4th set of three columns represents the non-linear component of the transformation (1/millimeters)
Column #10 > xy
Column #11 > xz
Column #12 > yz
The final 2 columns represent the quadratic terms of the eddy distortion correction (1/millimeters)
Column #13 > x2 - y2
Column #14 > 2z2 - x2 - y2
For details on the eddy distortion correction applied, please see; G.K. Rohde, A.S. Barnet, P.J. Basser, S. Marenco, C. Pierpaoli, Comprehensive approach for motion and distortion correction in diffusion weighted MRI. Magnetic Resonance in Medicine, vol. 51, pp. 103-114, 2004.
DMC.transformations
This file shows the rigid transformation of registering the 1st b0 image to the structural target image, and as such, there are values only in the first 6 columns of the first row. For coding reasons, there will be the number '1' in the 7th or 8th column depending on the phase encode direction, the 7th for a horizontal PE or the 8th for a vertical PE.
All subsequent lines will contain zeros, as the transformation for the 1st b0 volume is applied to all subsequent lines as described above, and the rigid transformation is calculated from the registration of the 1st b0 volume to the structural target image.
Source Code
If you have access to the source code, the following are relevant files
apply_trans_dwi.pro
apply_trans_t2.pro
cst_eval_mecc3d.pro
get_epi_coords_from_transformations.pro
